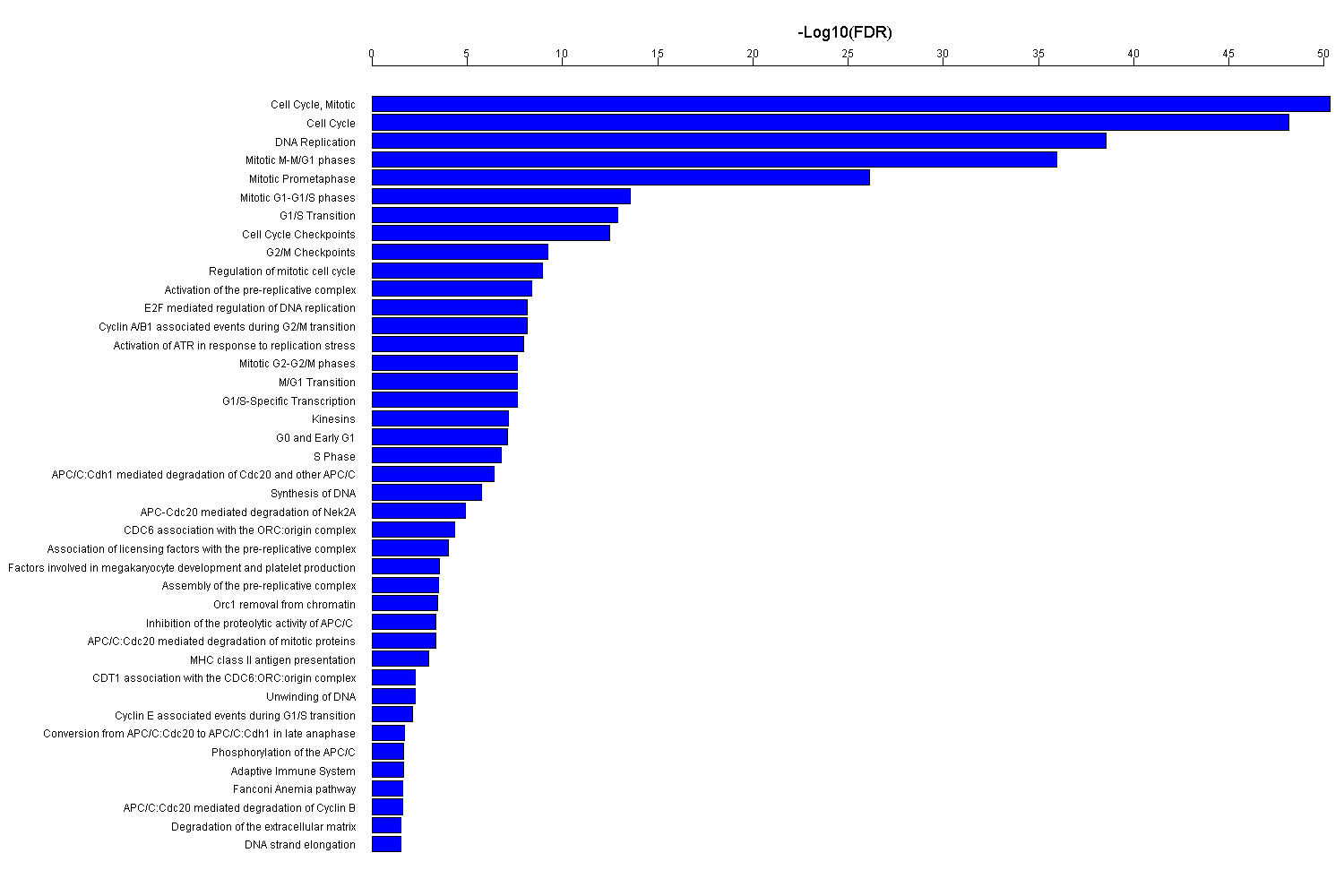
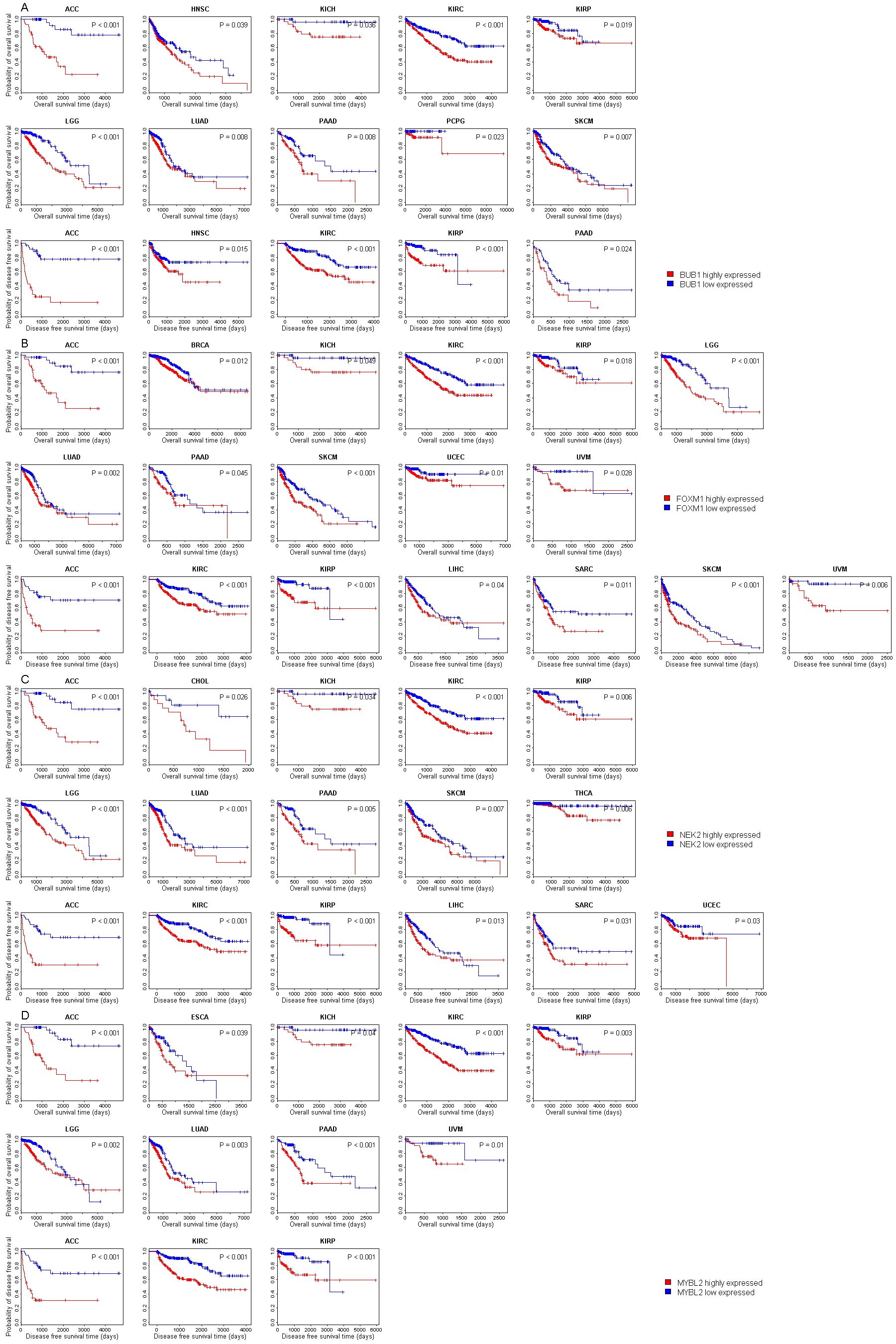
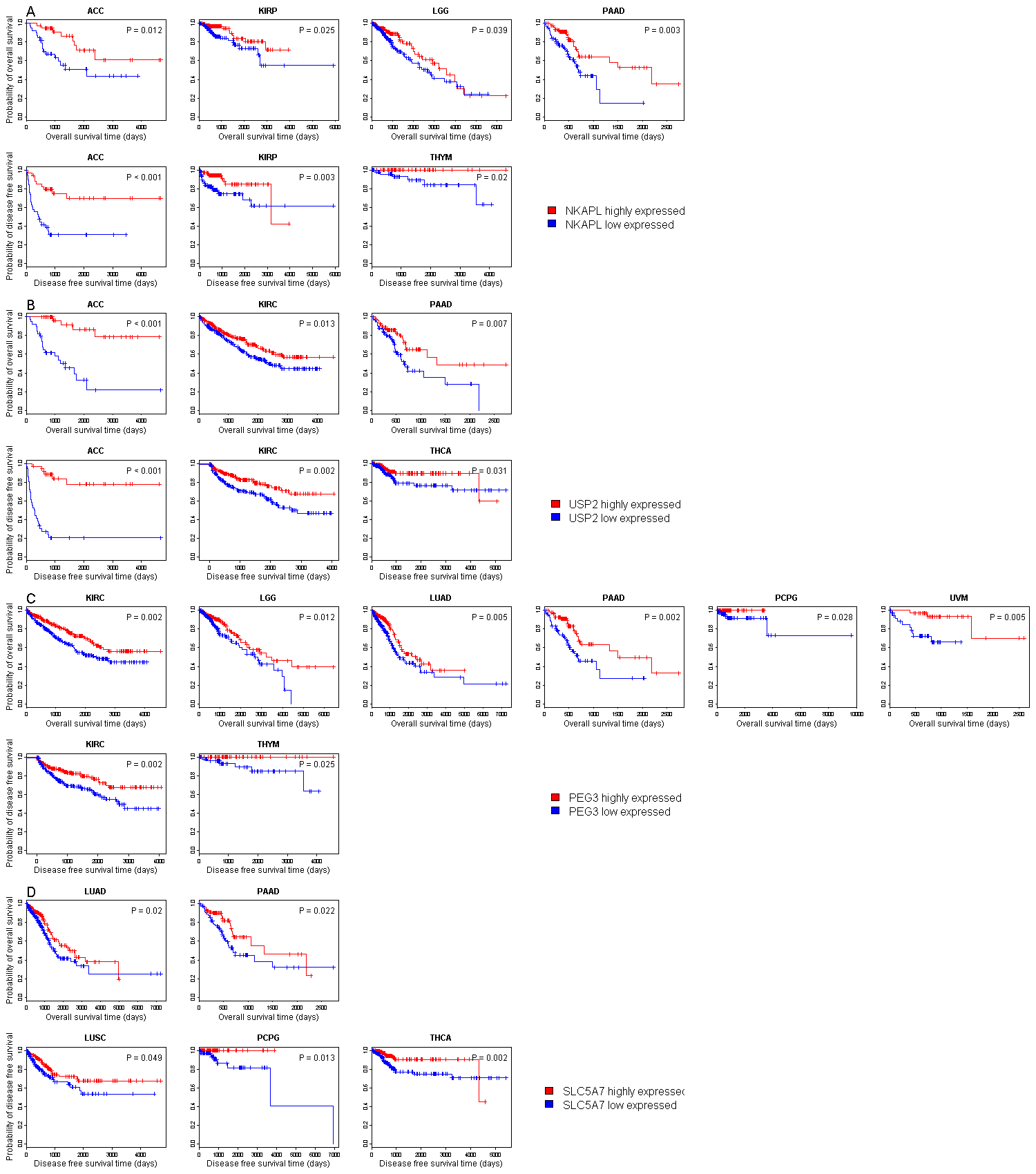
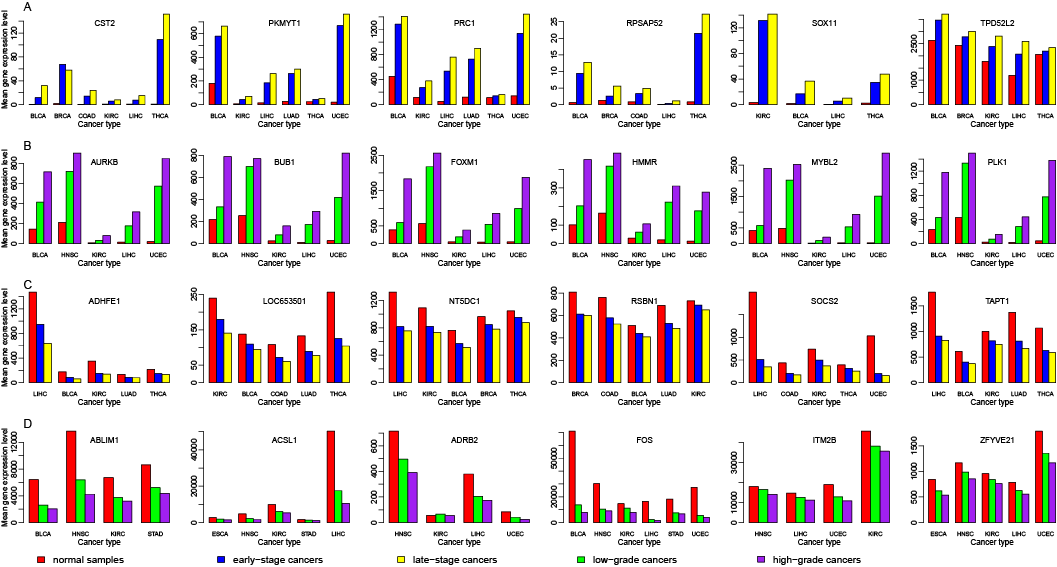
| Symbol |
Name |
Chromosome |
Cytoband |
| FLNC |
filamin C, gamma |
7 |
7q32-q35 |
| MYBL2 |
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
20 |
20q13.1 |
| CENPA |
centromere protein A |
2 |
2p23.3 |
| TPX2 |
TPX2, microtubule-associated |
20 |
20q11.2 |
| FOXM1 |
forkhead box M1 |
12 |
12p13 |
| RAB42 |
RAB42, member RAS oncogene family |
1 |
1p35.3 |
| CEP55 |
centrosomal protein 55kDa |
10 |
10q23.33 |
| KIF15 |
kinesin family member 15 |
3 |
3p21.31 |
| BUB1 |
BUB1 mitotic checkpoint serine/threonine kinase |
2 |
2q14 |
| BIRC5 |
baculoviral IAP repeat containing 5 |
17 |
17q25 |
| PLK1 |
polo-like kinase 1 |
16 |
16p12.2 |
| ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
1 |
1q31 |
| RRM2 |
ribonucleotide reductase M2 |
2 |
2p25-p24 |
| CDC20 |
cell division cycle 20 |
1 |
1p34.1 |
| NEIL3 |
nei endonuclease VIII-like 3 (E. coli) |
4 |
4q34.3 |
| NUF2 |
NUF2, NDC80 kinetochore complex component |
1 |
1q23.3 |
| KIF18B |
kinesin family member 18B |
17 |
17q21.31 |
| KIF14 |
kinesin family member 14 |
1 |
1q32.1 |
| TROAP |
trophinin associated protein |
12 |
12q13.12 |
| CDCA3 |
In multiple Geneids |
|
|
| UBE2C |
ubiquitin-conjugating enzyme E2C |
20 |
20q13.12 |
| IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
7 |
7p11 |
| MFI2 |
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
3 |
3q28-q29 |
| REEP2 |
receptor accessory protein 2 |
5 |
5q31 |
| GTSE1 |
G-2 and S-phase expressed 1 |
22 |
22q13.2-q13.3 |
| LAG3 |
lymphocyte-activation gene 3 |
12 |
12p13.32 |
| IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
3 |
3q27.2 |
| HMMR |
hyaluronan-mediated motility receptor (RHAMM) |
5 |
5q34 |
| CDC25C |
cell division cycle 25C |
5 |
5q31 |
| KIF20A |
kinesin family member 20A |
5 |
5q31 |
| FAM64A |
family with sequence similarity 64, member A |
17 |
17p13.2 |
| AURKB |
aurora kinase B |
17 |
17p13.1 |
| PDCD1 |
programmed cell death 1 |
2 |
2q37.3 |
| IQGAP3 |
IQ motif containing GTPase activating protein 3 |
1 |
1q21.3 |
| ZIC2 |
Zic family member 2 |
13 |
13q32 |
| PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
1 |
1q32.1 |
| MIR155HG |
MIR155 host gene |
21 |
|
| HJURP |
Holliday junction recognition protein |
2 |
2q37.1 |
| RDM1 |
RAD52 motif containing 1 |
17 |
17q11.2 |
| MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
19 |
19q13.1-q13.4 |
| C5orf34 |
chromosome 5 open reading frame 34 |
5 |
5p12 |
| SIT1 |
signaling threshold regulating transmembrane adaptor 1 |
9 |
9p13-p12 |
| EPHA6 |
In multiple Geneids |
|
|
| PTTG1 |
pituitary tumor-transforming 1 |
5 |
5q35.1 |
| CXCR3 |
In multiple Geneids |
|
|
| GZMK |
granzyme K (granzyme 3; tryptase II) |
5 |
5q11.2 |
| FAM26F |
family with sequence similarity 26, member F |
6 |
6q22.1 |
| CCDC8 |
coiled-coil domain containing 8 |
19 |
19q13.32 |
| SIX2 |
SIX homeobox 2 |
2 |
2p21 |
| CDCA5 |
In multiple Geneids |
|
|
| MCM10 |
minichromosome maintenance complex component 10 |
10 |
10p13 |
| KIF4A |
kinesin family member 4A |
X |
Xq13.1 |
| SHOX2 |
short stature homeobox 2 |
3 |
3q25.32 |
| MELK |
maternal embryonic leucine zipper kinase |
9 |
9p13.2 |
| MCM2 |
minichromosome maintenance complex component 2 |
3 |
3q21 |
| ECT2 |
epithelial cell transforming 2 |
3 |
3q26.1-q26.2 |
| CHEK1 |
checkpoint kinase 1 |
11 |
11q24.2 |
| APOE |
apolipoprotein E |
19 |
19q13.2 |
| ORC1 |
origin recognition complex, subunit 1 |
1 |
1p32 |
| BEND6 |
BEN domain containing 6 |
6 |
6p12.1 |
| GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
5 |
5q34-q35 |
| NCAPH |
non-SMC condensin I complex, subunit H |
2 |
2q11.2 |
| CENPI |
centromere protein I |
X |
Xq22.1 |
| DEPDC1 |
DEP domain containing 1 |
1 |
1p31.2 |
| KIF2C |
kinesin family member 2C |
1 |
1p34.1 |
| CDKN3 |
cyclin-dependent kinase inhibitor 3 |
14 |
14q22 |
| FAM72B |
In multiple Geneids |
|
|
| GINS1 |
GINS complex subunit 1 (Psf1 homolog) |
20 |
20p11.21 |
| RAD51AP1 |
RAD51 associated protein 1 |
12 |
12p13.2-p13.1 |
| CKAP2L |
cytoskeleton associated protein 2-like |
2 |
2q13 |
| FAM72D |
family with sequence similarity 72, member D |
1 |
1q21.1 |
| CDCA8 |
In multiple Geneids |
|
|
| SPHK1 |
sphingosine kinase 1 |
17 |
17q25.2 |
| EXO1 |
exonuclease 1 |
1 |
1q43 |
| DLGAP5 |
discs, large (Drosophila) homolog-associated protein 5 |
14 |
14q22.3 |
| KIF23 |
kinesin family member 23 |
15 |
15q23 |
| DEPDC1B |
DEP domain containing 1B |
5 |
5q12.1 |
| SUSD3 |
sushi domain containing 3 |
9 |
9q22.31 |
| NEK2 |
NIMA-related kinase 2 |
1 |
1q32.3 |
| DTL |
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
1 |
1q32 |
| FANCB |
Fanconi anemia, complementation group B |
X |
Xp22.2 |
| NDC80 |
NDC80 kinetochore complex component |
18 |
18p11.32 |
| CCNA2 |
cyclin A2 |
4 |
4q27 |
| PRC1 |
protein regulator of cytokinesis 1 |
15 |
15q26.1 |
| KIF11 |
kinesin family member 11 |
10 |
10q24.1 |
| CENPF |
centromere protein F, 350/400kDa |
1 |
1q41 |
| CCNB2 |
cyclin B2 |
15 |
15q22.2 |
| PRR11 |
proline rich 11 |
17 |
17q22 |
| ALPK2 |
alpha-kinase 2 |
18 |
18q21.31 |
| CLSPN |
claspin |
1 |
1p34.2 |
| GAS1 |
growth arrest-specific 1 |
9 |
9q21.3-q22 |
| KIF18A |
kinesin family member 18A |
11 |
11p14.1 |
| GSG2 |
germ cell associated 2 (haspin) |
17 |
17p13 |
| TCF19 |
transcription factor 19 |
6 |
6p21.3 |
| OIP5 |
Opa interacting protein 5 |
15 |
15q15.1 |
| SPC25 |
SPC25, NDC80 kinetochore complex component |
2 |
2q31.1 |
| FAM20A |
family with sequence similarity 20, member A |
17 |
17q24.2 |
| MKI67 |
marker of proliferation Ki-67 |
10 |
10q26.2 |
| SGOL2 |
shugoshin-like 2 (S. pombe) |
2 |
2q33.1 |
| NCAPG |
non-SMC condensin I complex, subunit G |
4 |
4p15.33 |
| SGOL1 |
shugoshin-like 1 (S. pombe) |
3 |
3p24.3 |
| RAD54L |
RAD54-like (S. cerevisiae) |
1 |
1p32 |
| WDR62 |
WD repeat domain 62 |
19 |
19q13.12 |
| RAD51 |
RAD51 recombinase |
15 |
15q15.1 |
| E2F1 |
E2F transcription factor 1 |
20 |
20q11.2 |
| TENM3 |
teneurin transmembrane protein 3 |
4 |
4q35.1 |
| C1R |
complement component 1, r subcomponent |
12 |
12p13 |
| SLAMF8 |
SLAM family member 8 |
1 |
1q23.2 |
| EPR1 |
effector cell peptidase receptor 1 (non-protein coding) |
17 |
17q25 |
| FAM72A |
In multiple Geneids |
|
|
| NUSAP1 |
nucleolar and spindle associated protein 1 |
15 |
15q15.1 |
| SKA3 |
spindle and kinetochore associated complex subunit 3 |
13 |
13q12.11 |
| ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
8 |
8p21.1 |
| TRIP13 |
thyroid hormone receptor interactor 13 |
5 |
5p15.33 |
| XRCC2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
7 |
7q36.1 |
| KIFC1 |
kinesin family member C1 |
6 |
6p21.3 |
| UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
4 |
4p14 |
| CDC45 |
cell division cycle 45 |
22 |
22q11.21 |
| PKMYT1 |
protein kinase, membrane associated tyrosine/threonine 1 |
16 |
16p13.3 |
| SIGLEC1 |
sialic acid binding Ig-like lectin 1, sialoadhesin |
20 |
20p13 |
| POLQ |
polymerase (DNA directed), theta |
3 |
3q13.33 |
| TYMS |
thymidylate synthetase |
18 |
18p11.32 |
| BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
15 |
15q15 |
| TOP2A |
topoisomerase (DNA) II alpha 170kDa |
17 |
17q21-q22 |
| SKA1 |
spindle and kinetochore associated complex subunit 1 |
18 |
18q21.1 |
| CENPE |
centromere protein E, 312kDa |
4 |
4q24-q25 |
| ARHGAP11A |
Rho GTPase activating protein 11A |
15 |
15q13.2 |
| ERCC6L |
excision repair cross-complementation group 6-like |
X |
Xq13.1 |
| PDGFRL |
platelet-derived growth factor receptor-like |
8 |
8p22-p21.3 |
| CCNB1 |
cyclin B1 |
5 |
5q12 |
| ARHGAP29 |
Rho GTPase activating protein 29 |
1 |
1p22.1 |
| PIF1 |
In multiple Geneids |
|
|
| CDCA2 |
In multiple Geneids |
|
|
| 3-Sep |
Data not found |
|
|
| ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
12 |
12q |
| CHRDL2 |
chordin-like 2 |
11 |
11q14 |
| SALL4 |
spalt-like transcription factor 4 |
20 |
20q13.2 |
| GINS4 |
GINS complex subunit 4 (Sld5 homolog) |
8 |
8p11.21 |
| CXCL13 |
chemokine (C-X-C motif) ligand 13 |
4 |
4q21 |
| STAC |
SH3 and cysteine rich domain |
3 |
3p22.3 |
| CDCA7 |
cell division cycle associated 7 |
2 |
2q31 |
| GBP5 |
guanylate binding protein 5 |
1 |
1p22.2 |
| C17orf53 |
chromosome 17 open reading frame 53 |
17 |
17q21.31 |
| TTK |
TTK protein kinase |
6 |
6q14.1 |
| ADAMDEC1 |
ADAM-like, decysin 1 |
8 |
8p21.2 |
| ASF1B |
anti-silencing function 1B histone chaperone |
19 |
19p13.12 |
| C2orf48 |
chromosome 2 open reading frame 48 |
2 |
2p25.1 |
| FXYD6 |
FXYD domain containing ion transport regulator 6 |
11 |
11q23.3 |
| E2F2 |
E2F transcription factor 2 |
1 |
1p36 |
| NECAB1 |
N-terminal EF-hand calcium binding protein 1 |
8 |
8q21.3 |
| HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
6 |
6p21.3 |
| RAB39B |
RAB39B, member RAS oncogene family |
X |
Xq28 |
| COL8A1 |
collagen, type VIII, alpha 1 |
3 |
3q12.3 |
| ARHGAP11B |
In multiple Geneids |
|
|
| PBK |
PDZ binding kinase |
8 |
8p21.2 |
| CDC25A |
cell division cycle 25A |
3 |
3p21 |
| CPVL |
carboxypeptidase, vitellogenic-like |
7 |
7p15.1 |
| CLGN |
calmegin |
4 |
4q28.3-q31.1 |
| IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
17 |
17q21.32 |
| HLA-DQA2 |
major histocompatibility complex, class II, DQ alpha 2 |
6 |
6p21.3 |
| KREMEN2 |
kringle containing transmembrane protein 2 |
16 |
16p13.3 |
| CD19 |
CD19 molecule |
16 |
16p11.2 |
| CXCR6 |
chemokine (C-X-C motif) receptor 6 |
3 |
3p21 |
| CXCL9 |
chemokine (C-X-C motif) ligand 9 |
4 |
4q21 |
| LTA |
lymphotoxin alpha |
6 |
6p21.3 |
| CD72 |
CD72 molecule |
9 |
9p13.3 |
| CDKN2C |
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
1 |
1p32 |
| CXCL10 |
chemokine (C-X-C motif) ligand 10 |
4 |
4q21 |
| FNDC5 |
fibronectin type III domain containing 5 |
1 |
1p35.1 |
| MEI1 |
meiosis inhibitor 1 |
22 |
22q13.2 |
| LOC90768 |
uncharacterized LOC90768 |
4 |
4q34.3 |
| MTFR2 |
mitochondrial fission regulator 2 |
6 |
6q23.3 |
| GBP1 |
guanylate binding protein 1, interferon-inducible |
1 |
1p22.2 |
| CR2 |
complement component (3d/Epstein Barr virus) receptor 2 |
1 |
1q32 |
| ELOVL2 |
ELOVL fatty acid elongase 2 |
6 |
6p24.2 |
| CD2 |
CD2 molecule |
1 |
1p13.1 |
| HMGA2 |
high mobility group AT-hook 2 |
12 |
12q15 |
| CHRDL1 |
chordin-like 1 |
X |
Xq23 |
| CCL5 |
chemokine (C-C motif) ligand 5 |
17 |
17q12 |
| RMI2 |
RecQ mediated genome instability 2 |
16 |
16p13.13 |
| DES |
desmin |
2 |
2q35 |
| FDCSP |
follicular dendritic cell secreted protein |
4 |
4q13 |
| HOXB13 |
homeobox B13 |
17 |
17q21.2 |
| HAMP |
hepcidin antimicrobial peptide |
19 |
19q13.1 |
| CCL19 |
chemokine (C-C motif) ligand 19 |
9 |
9p13 |
| SPIB |
Spi-B transcription factor (Spi-1/PU.1 related) |
19 |
19q13.3-q13.4 |
| CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
11 |
11q23 |
| FOXD3 |
forkhead box D3 |
1 |
1p31.3 |
| C3 |
complement component 3 |
19 |
19p13.3-p13.2 |
| LRP8 |
In multiple Geneids |
|
|
| SMC1B |
structural maintenance of chromosomes 1B |
22 |
22q13.31 |
| CXCR2P1 |
chemokine (C-X-C motif) receptor 2 pseudogene 1 |
2 |
2q35 |
| PLA2G2D |
phospholipase A2, group IID |
1 |
1p36.12 |
| UBASH3A |
ubiquitin associated and SH3 domain containing A |
21 |
21q22.3 |
| MAP1LC3C |
microtubule-associated protein 1 light chain 3 gamma |
1 |
1q43 |
| ADAM6 |
ADAM metallopeptidase domain 6 (pseudogene) |
14 |
14q32.33 |
| SLC2A4 |
solute carrier family 2 (facilitated glucose transporter), member 4 |
17 |
17p13 |
| CHI3L2 |
chitinase 3-like 2 |
1 |
1p13.3 |
| GBP1P1 |
guanylate binding protein 1, interferon-inducible pseudogene 1 |
1 |
1p22.2 |
| IL2RG |
interleukin 2 receptor, gamma |
X |
Xq13.1 |
| LTB |
lymphotoxin beta (TNF superfamily, member 3) |
6 |
6p21.3 |
| GDF1 |
growth differentiation factor 1 |
19 |
19p12 |
| LOC399815 |
chromosome 10 open reading frame 88 pseudogene |
10 |
10q26.13 |
| HLA-DOA |
major histocompatibility complex, class II, DO alpha |
6 |
6p21.3 |
| JAK3 |
Janus kinase 3 |
19 |
19p13.1 |
| ZAP70 |
zeta-chain (TCR) associated protein kinase 70kDa |
2 |
2q12 |
| CD8B |
CD8b molecule |
2 |
2p12 |
| ZNF683 |
zinc finger protein 683 |
1 |
1p36.11 |
| NTNG1 |
In multiple Geneids |
|
|
| IDO1 |
indoleamine 2,3-dioxygenase 1 |
8 |
8p12-p11 |
| TNNT1 |
troponin T type 1 (skeletal, slow) |
19 |
19q13.4 |
| LGALS17A |
Charcot-Leyden crystal protein pseudogene |
19 |
19q13.2 |
| FABP4 |
fatty acid binding protein 4, adipocyte |
8 |
8q21 |
| VIPR1 |
vasoactive intestinal peptide receptor 1 |
3 |
3p22 |
| GPX2 |
glutathione peroxidase 2 |
14 |
14q24.1 |
| DUOX1 |
dual oxidase 1 |
15 |
15q15.3 |
| LAD1 |
ladinin 1 |
1 |
1q25.1-q32.3 |
| TMEM184A |
transmembrane protein 184A |
7 |
7p22.3 |
| OVOL1 |
ovo-like zinc finger 1 |
11 |
11q13 |
| FUT3 |
fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) |
19 |
19p13.3 |
| FAM3D |
family with sequence similarity 3, member D |
3 |
3p14.2 |
| RAPGEFL1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
17 |
17q21.1 |
| AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
7 |
7q22.1 |
| DEGS2 |
delta(4)-desaturase, sphingolipid 2 |
14 |
14q32.2 |
| ALDH1L1 |
aldehyde dehydrogenase 1 family, member L1 |
3 |
3q21.3 |
| C10orf99 |
chromosome 10 open reading frame 99 |
10 |
10q23.1 |
| TCN1 |
transcobalamin I (vitamin B12 binding protein, R binder family) |
11 |
11q11-q12 |
| TMPRSS4 |
transmembrane protease, serine 4 |
11 |
11q23.3 |
| ZBTB7C |
In multiple Geneids |
|
|
| SLC44A4 |
solute carrier family 44, member 4 |
6 |
6p21.3 |
| BCAS1 |
breast carcinoma amplified sequence 1 |
20 |
20q13.2 |
| FSTL4 |
follistatin-like 4 |
5 |
5q31.1 |
| ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
16 |
16q24.1 |
| CA12 |
carbonic anhydrase XII |
15 |
15q22 |
| TMEM154 |
transmembrane protein 154 |
4 |
4q31.3 |
| CYP2J2 |
cytochrome P450, family 2, subfamily J, polypeptide 2 |
1 |
1p31.3-p31.2 |
| CNGA1 |
cyclic nucleotide gated channel alpha 1 |
4 |
4p12 |
| DUOXA1 |
dual oxidase maturation factor 1 |
15 |
15q21.1 |
| CCDC64B |
coiled-coil domain containing 64B |
16 |
16p13.3 |
| IYD |
iodotyrosine deiodinase |
6 |
6q25.1 |
| CNTN3 |
contactin 3 (plasmacytoma associated) |
3 |
3p12.3 |
| GOLGA7B |
golgin A7 family, member B |
10 |
10q24.2 |
| PTPRR |
protein tyrosine phosphatase, receptor type, R |
12 |
12q15 |
| MSX2 |
msh homeobox 2 |
5 |
5q35.2 |
| REEP6 |
receptor accessory protein 6 |
19 |
19p13.3 |
| SMIM5 |
small integral membrane protein 5 |
17 |
17q25.1 |
| EVPL |
envoplakin |
17 |
17q25 |
| WIF1 |
WNT inhibitory factor 1 |
12 |
12q14.3 |
| GJB5 |
gap junction protein, beta 5, 31.1kDa |
1 |
1p35.1 |
| SULT2B1 |
sulfotransferase family, cytosolic, 2B, member 1 |
19 |
19q13.3 |
| CEACAM5 |
carcinoembryonic antigen-related cell adhesion molecule 5 |
19 |
19q13.1-q13.2 |
| TGM3 |
transglutaminase 3 |
20 |
20q11.2 |
| TMPRSS2 |
transmembrane protease, serine 2 |
21 |
21q22.3 |
| DSC2 |
desmocollin 2 |
18 |
18q12.1 |
| CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
19 |
19p13.2 |
| SPINK5 |
serine peptidase inhibitor, Kazal type 5 |
5 |
5q32 |
| PVRL4 |
poliovirus receptor-related 4 |
1 |
1q23.3 |
| CEACAM6 |
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
19 |
19q13.2 |
| UNC93A |
unc-93 homolog A (C. elegans) |
6 |
6q27 |
| CALML3 |
calmodulin-like 3 |
10 |
10p15.1 |
| FGFBP1 |
fibroblast growth factor binding protein 1 |
4 |
4p15.32 |
| KRT31 |
keratin 31, type I |
17 |
17q21.2 |
| SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
22 |
22q12.3 |
| HOMER2 |
homer scaffolding protein 2 |
15 |
15q24.3 |
| CYP39A1 |
cytochrome P450, family 39, subfamily A, polypeptide 1 |
6 |
6p12.3 |
| TMEM125 |
transmembrane protein 125 |
1 |
1p34.2 |
| HKDC1 |
In multiple Geneids |
|
|
| SOWAHB |
sosondowah ankyrin repeat domain family member B |
4 |
4q21.1 |
| FMO2 |
flavin containing monooxygenase 2 (non-functional) |
1 |
1q24.3 |
| CRYM |
crystallin, mu |
16 |
16p12.2 |